Examples of SWEEP in use
Smart sensing in agriculture
Sensors that provide real-time data from the fields can give valuable insights into physiological traits of plants, allowing for optimization of resources and promoting smarter farming practices. This can be of tremendous value for farmers, but the fine temporal resolution of the data leads to large volumes that can be challenging to maintain and analyze.
SWEEP powers the agri-sensing platform plantbeats.io, helping provide quick and intuitive summarization of large amounts of sensor data through scalable cloud-based workflows.

Satellite imagery analysis
Satellite images are an important source of information about the Earth’s surface, and are widely used to detect and monitor the effects of climate change on our environment. Many organizations make imagery data available via web services, often as numerous small image segments that need to be individually downloaded and analyzed via image-processing and machine learning pipelines, after which the results are gathered and compiled together.
Such data pipelines are suitable for running as microservices-style workflows, consisting of many small tasks that can be run independently. SWEEP has been used to build and execute several Function-as-a-Service-based satellite imagery workflows.
In this project , the color of remote Arctic lakes was analyzed in order to get an indication of the effects of a warming climate on ecosystems. In another project, remote sensing was used to monitor the effects of climate change on wildflower communities in a national park.
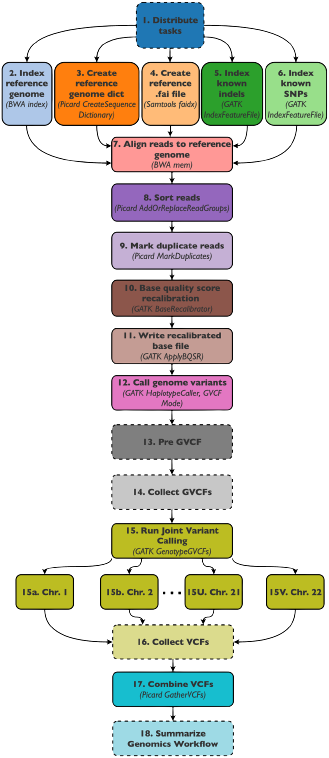
Next-generation sequencing
Advances in sequencing technologies during recent years have revolutionized genomic research, revealing new insights into our evolutionary past as well as improving medical diagnosis and paving way for optimized strategies such as precision medicine and personalized diagnosis.
Genomic data analysis pipelines are demanding because they tend to be computationally and data intensive, posing several challenges to the institutes where they are run. Traditionally they have required access to dedicated high-performance resources, such as large-scale computing clusters, but the increased adoption of cloud computing is aiding in making the resources required for analysis of NGS data more easily available.
In many cases, the move to a distributed, cloud-based execution model requires some modification of the batch-style processing format that genomic pipelines typically have. SWEEP has been used to redesign a classical variant-calling workflow and to execute it in a multi-cloud environment using serverless resources of AWS and Microsoft Azure.